Category Archives: Chemometrics
EigenU Registrations Coming In!
Jan 23, 2012
Registrations have started coming in for Eigenvector University 2012. This seventh annual EigenU will be May 13-18 at the Washington Athletic Club in Seattle.
New for this year, Batch Multivariate Statistical Process Control for PAT combines the technical aspects of developing chemometric models for monitoring batch processes with the practical aspects of implementing and deploying models, particularly in the pharmaceutical industries. Our DOE course, which debuted last year, has been updated and expanded to become Design of Experiments for QbD (Quality by Design). Also updated this year, Advanced Preprocessing for Spectral Applications has been refocused on spectroscopy.
The PLS_Toolbox/Solo User Poster Session returns with Apple iPod prizes for the two best posters. New and advanced features of our software will be highlighted in the PowerUser Tips & Tricks evening session. And of course our traditional group dinner will be held at Torchy’s in the WAC.
Our most popular classes usually fill up, so register early! Discount registration rates apply for registrations received with payment by April 11, 2012.
See you in Seattle!
BMW
PLS_Toolbox in Research and Publications
Dec 6, 2011
Our Chief of Technology Development Jeremy M. Shaver received a very nice letter this morning from Balázs Vajna, who is a Ph.D. student at Budapest University of Technology and Economics. As you’ll see from the references below, he is a very productive young man! Here is his letter to Jeremy, highlighting how he used PLS_Toolbox in his work:
Dear Jeremy,
I would like to thank you for all your help with the Eigenvector products. With your help, I was able to successfully carry out detailed investigations using chemical imaging and chemometric evaluation in such a way that I could publish these results in relevant international journals. I would like to draw your attention to the following publications where (only) PLS_Toolbox was used for chemometric evaluation:
- B. Vajna, I. Farkas, A. Farkas, H. Pataki, Zs. Nagy, J. Madarász, Gy. Marosi, “Characterization of drug-cyclodextrin formulations using Raman mapping and multivariate curve resolution,” Journal of Pharmaceutical and Biomedical Analysis, 56, 38-44, 2011.
- B. Vajna, H. Pataki, Zs. Nagy, I. Farkas, Gy. Marosi, “Characterization of melt extruded and conventional Isoptin formulations using Raman chemical imaging and chemometrics,” International Journal of Pharmaceutics, 419, 107-113, 2011.
These may be considered as showcases of using PLS_Toolbox in Raman chemical imaging, and – which is maybe even more interesting in the light of your collaboration with Horiba Jobin Yvon – the joint use of PLS_Toolbox and LabSpec. The following studies have also been published where MCR-ALS and SMMA (Purity) were carried out with PLS_Toolbox and were tested along with other curve resolution techniques.
- B. Vajna, G. Patyi, Zs. Nagy, A. Farkas, Gy. Marosi, “Comparison of chemometric methods in the analysis of pharmaceuticals with hyperspectral Raman imaging,” Journal of Raman Spectroscopy, 42(11), 1977-1986, 2011.
- B. Vajna, A. Farkas, H. Pataki, Zs. Zsigmond, T. Igricz, Gy. Marosi, “Testing the performance of pure spectrum resolution from Raman hyperspectral images of differently manufactured pharmaceutical tablets,” Analytica Chimica Acta, in press.
- B. Vajna, B. Bodzay, A. Toldy, I. Farkas, T. Igricz, G. Marosi, “Analysis of car shredder polymer waste with Raman mapping and chemometrics,” Express Polymer Letters, 6(2), 107-119, 2012.
I just wanted to let you know that these publications exist, all using PLS_Toolbox in the evaluaton of Raman images, and that I am very grateful for your help throughout. I hope you will find them interesting.
Best regards,
Balázs
—
Balázs Vajna
PhD student
Department of Organic Chemistry and Technology
Budapest University of Technology and Economics
8 Budafoki str., H-1111 Budapest, Hungary
Thanks, Balázs, your letter just made our day! We’re glad you found our tools useful!
BMW
Missing Data (part three)
Nov 21, 2011
In the first and second installments of this series, we considered aspects of using an existing PCA model to replace missing variables. In this third part, we’ll move on to using PLS models.
Although it was shown previously that PCA can be used to perfectly impute missing values in rank deficient, noise free data, it’s not hard to guess that PCA might be suboptimal with regards to imputing missing elements in real, noisy data. The goal of PCA, after all, is to estimate the data subspace, not predict particular elements. Prediction is typically the goal of regression methods, such as Partial Least Squares. In fact, regression models can be used to construct estimates of any and all variables in a data set based on the remaining variables. In our 1989 AIChE paper we proposed comparing those estimates to actual values for the purpose of fault detection. Later this became known as regression adjusted variables, as in Hawkins, 1991.
There is a little known function in PLS_Toolbox, (since the first version in 1989 or 90), plsrsgn, that can be used to develop collections of PLS models, where each variable in a data set is predicted by the remaining variables. The regression vectors are mapped into a matrix that generates the residuals between the actual and predicted values in much the same way as the I–PP‘ matrix from PCA.

We can compare the results of using these collections of PLS models to using the PCA done previously. Here we created the coeff matrix using (a conservative) 3 LVs in each of the PLS submodels. Each sub model could of course be optimized individually, but for illustration purposes this will be adequate. The reconstruction error of the PLS models is compared with PCA in the figure shown at left, where the error for the collection of PLS models is shown in red, superimposed over the reconstruction via the PCA model error, in blue. The PLS models’ error is lower for each variable, in some cases, substantially, e.g. variables 3-5.
![]()
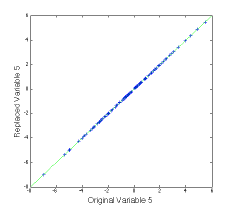
The second figure, at left, shows the estimate of variable 5 for both the PLS (green) and PCA (red) methods compared to the measured values (blue). It is clear that the PLS model tracks the actual value much better.
Because the estimation error is smaller, collections of PLS models can be much more sensitive to process faults than PCA models, particularly individual sensor faults.
It is also possible to replace missing variables based on these collections of PLS models in (nearly) exactly the same manner as in PCA. The difference is that, unlike in PCA, the matrix which generates the residuals is not symmetric, so the R12 term (see part one) does not equal R21‘. The solution is to calculate b using their average, thus
b = 0.5(R12 + R21‘)R11-1
Curiously, unlike the PCA case, the residuals on the replaced variables will not be zero except in the unlikely case that R12 = R21‘.
In the case of an existing single PLS model, it is of course possible to use this methodology to estimate the values of missing variables based on the PLS loadings. (Or, if you insist, on the PLS weights. Given that residuals based on weights are larger than residuals based on loadings, I’d expect better luck reconstructing from the loadings but I offer that here without proof.)
In the next installment of this series, we will consider the more challenging problem of building models on incomplete data records.
BMW
B.M. Wise, N.L. Ricker, and D.J. Veltkamp, “Upset and Sensor Failure Detection in Multivariate Pocesses,” AIChE Annual Meeting, 1989.
D.M. Hawkins, “Multivariate Quality Control Based on Regression Adjusted Variables,” Technometrics, Vol. 33, No. 1, 1991.
2011 EAS Awards for NIR and Chemometrics
Nov 17, 2011
I had the privilege of being involved with two award sessions at this week’s Eastern Analytical Symposium (EAS). I was very pleased to be invited to speak in the session honoring former Eigenvectorian Charles E. “Chuck” Miller for Outstanding Achievements in Near Infrared Spectroscopy. Chuck elected to have talks from speakers that represented phases in his career. This included Robert Thompson, Chuck’s advisor from Oberlin College, Tormod Næs, from his time at University of Washington’s Center for Process Analytical Chemistry (CPAC) and Matforsk (now Nofima), Cary Sohl of DuPont, and myself. Chuck, now with Merck, presented “26 Years of NIR Technology – From One Person’s Perspective,” which chronicled his career and influences, along with the progression of NIR over the period.
The session was organized by Katherine Bakeev of CAMO. Pictured below are Katherine, Tormod, Cary, Chuck, EAS President David Russell, and myself.

The session provided ample evidence of the intertwined evolution of chemometrics and NIR, with two primarily chemometric talks and two NIR talks with aspects of chemometrics.
I was also our representative at the session honoring Beata Walczak of the University of Silesian in Poland. Beata was the recipient of the EAS Award for Outstanding Achievements in Chemometrics, sponsored once again by Eigenvector Research. Beata and I are pictured below with the award.

The award session, organized by Peter D. Wentzell of Dalhousie University, had an “omics” theme with talks on metabolomics and proteomics. Speakers included Peter, Tobais Karakach of the Institute for Marine Biosciences, Sarah Rutan of Virginia Commonwealth University and Michal Daszykowski, also of Silesian. Beata presented “Chemometrics in Proteomics,” an overview of her work in the field highlighting methods for aligning samples from 2-D gel electrophoresis.
Congratulations to both Chuck and Beata on two very well-deserved awards!
BMW
Missing Data (part two)
Nov 11, 2011
In Missing Data (part one) I outlined an approach for in-filling missing data when applying an existing Principal Components Analysis (PCA) model. Let us now consider when this approach might be expected to fail. Recall that missing data estimation results in a least-squares problem with solution:
xb = –xgR21R11-1
In our short courses, I advise students to be wary any time a matrix inverse is used, and this case is no exception. Inverses are defined only for matrices of full rank, and may be unstable for nearly rank-deficient matrices. So under what conditions might we expect R11 to be rank deficient? Recall that R11 is the part of I–PP‘ that applies to the variables which we want to replace. Problems arise when the variables to be replaced form a group that are perfectly correlated with each other but not with any of the remaining variables. When this happens the variables will either be 1: included as a group in the PCA model (if enough PCs are retained) or 2: excluded as a group (too few PCs retained). In case 1, R11 is rank deficient and the inverse isn’t defined. In case 2, R11 is just I, but the loadings of the correlated group are zero, so the R12 part of the solution is 0. In either case, it makes sense that a solution isn’t possible–what information would it be based on?
With real data, of course, it is highly unlikely that R11 will be rank deficient to within numerical precision (or that R12 will be zero). But it certainly may happen that R11 is near rank deficient, in which case the estimates of the missing variables will not be very good. Fortunately, in most systems the measured variables are somewhat correlated with each other and the method can be employed.

In their 1995 paper, Nomikos and MacGregor estimated the value of missing variables using a truncated Classical Least Squares (CLS) formulation. The PCA loadings are fit to the available data, leaving out the missing portions, to estimate scores which are then used to estimate missing values. This reduces to:
xb = xg(PgPg‘)-1PgPb‘
where Pb and Pg refer to the part of the PCA model loadings for the missing (bad) and available (good) data, respectively. In 1996 Nelson, Taylor and MacGregor noted that this method was equivalent to the method in our 1991 paper but offered no proof. The proof can be found in “Refitting PCA, MPCA and PARAFAC Models to Incomplete Data Records” from FACSS, 2007.

So how does this work in practice? The topmost figure shows the estimation error for each of the 20 variables in the melter data based on a 4 PC models with mean-centering. The model was estimated with every other sample and tested on the other samples. The estimation error is shown in units of Relative Standard Deviation (RSD) to the raw data. Thus, the variables with error near 1.0 aren’t being predicted any better than just using the mean value, while the variables with error below 0.2 are tracking quite well. An example is shown in the middle figure, which shows temperature sensor number 8 actual (blue line) and predicted (red x) for the test set as a function sample number (time).

The reason for the large differences in ability to replace variables in this data set is, of course, directly related to how independent the variables are. A graphic illustration of this can be produced with the PLS_Toolbox corrmap function, which produced the third figure. The correlation matrix for the temperatures is colored red where there is high positive correlation, blue for negative correlation, and white for no correlation. It can be seen that variables with low estimation error (e.g. 7, 8, 17, 18) are strongly correlated with other variables, whereas variables with high estimation error (e.g. 2, 12) are not correlated strongly with any other variables.
To summarize, we’ve shown that missing variables can be imputed based on an existing PCA model and the available measurements. This success of this approach depends upon the degree to which the missing variables are correlated with available variables, as might be expected. In the next installment of this Missing Data series, we’ll explore using regression models, particularly Partial Least Squares (PLS) to replace missing data.
BMW
P. Nomikos and J.F. MacGregor, “Multivariate SPC Charts for Monitoring Batch Processes,” Technometrics, 37(1), pps. 41-58, 1995.
P.R.C. Nelson, P.A. Taylor and J.F. MacGregor, “Missing data method in PCA and PLS: Score calculations with incomplete observations,” Chemometrics & Intell. Lab. Sys., 35(1), pps. 45-65, 1996.
B.M. Wise, “Re-fitting PCA, MPCA and PARAFAC Models to Incomplete Data Records,” FACSS, Memphis, TN, October, 2007.
Missing Data (part one)
Nov 5, 2011
Over the next few weeks I’m going to be discussing some aspects of missing data. This is an important aspect of chemometrics as many applications suffer from this problem. Missing data is especially common in process applications where there are many independent sensors.
I got interested in missing data while in graduate school in the late 1980s. I worked a lot with a prototype glass melter for the solidification of nuclear fuel reprocessing waste. The primary measurements were temperatures provided by thermocouple sensors. The very high temperatures in this system, nearing 1200C (~2200F), caused the thermocouples to fail frequently. Thus it was common for the data record to be incomplete.
Missing data is also common in batch process monitoring. There are several approaches for building models on complete, finished batches. However, it is most useful to know if batches are going wrong BEFORE they are complete. Thus, it is desirable to be able to apply the model to an incomplete data record.
Missing data problems can be divided into two classes: 1)those involving missing data when applying an existing model to new data records, and 2) those involving building a model on an incomplete data record. Of these, the first problem is by far the easiest to deal with, so we will start with it. It will, however, illustrate some approaches which can be modified for use in the second case. These approaches can also be used for other purposes as well, such as cross-validation of Principal Component Analysis (PCA) models.
Consider now the case where you have a process that periodically produces a new data vector xi (1 x n). With it you have a validated PCA model, with loadings Pk (n x k). The residual sum-of-squares or Q statistic, can be calculated for the ith sample as Q = xiRxi‘ where R = I–PkPk‘. For the sake of convenience, imagine that the first p variables in this model are no longer available, but the remaining n–p variables are as usual. Thus, x can be partitioned into a group of bad variables xb and a group of good variables xg, x = [xb xg]. The calculation of Q can then be broken down into parts which do and do not involve missing variables:
Q = xbR11xb‘ + xgR21xb‘ + xbR12xg‘ + xgR22xg‘
where R11 is the upper left (p x p) part of R, R12 = R21‘ is the lower left (n–p x p) section, and R22 is the lower right (n–p x n–p) section.
It is possible to solve for the values of the bad variables xb that minimize Q, as shown in our 1991 paper referenced below. The (incredibly simple) solution is
xb = –xgR21R11-1
Unsurprisingly, the residuals on the replaced variables on the full model will be zero.
This method is the basis of the PLS_Toolbox function replace, which maps the solution above into a matrix so variables in arbitrary positions can be replaced.

It is easy to demonstrate that this method works perfectly in the rank deficient, no noise case. In MATLAB, you can create a rank 5 data set with 20 variables, then use the Singular Value Decomposition (SVD) to get a set of PCA loadings P, and from that, the R matrix.
>> c = randn(100,5);
>> p = randn(20,5);
>> x = c*p’;
>> [u,s,v] = svd(x);
>> P = v(:,1:5);
>> R = eye(20)-P*P’;
Now let’s say the sensor associated with variable 5 has failed. We can use the replace function to generate a matrix Rm which replaces it based on the values of the other variables.
>> Rm = replace(R,5,’matrix’);
>> imagesc(sign(Rm)), colormap(rwb)
Rm has the somewhat curious structure show in the figure above. The white area is zeros, the diagonal is ones, and R21R11-1 for the appropriately rearranged R is mapped into the vertical section.
We can try Rm out on a new data set that spans the same space as the previous one, and plot up the results as follows:
>> newx = randn(100,5)*p’;
>> var5 = newx(:,5);
>> newx(:,5) = 0;
>> newx_r = newx*Rm;
>> figure(2)
>> plot(var5,newx_r(:,5),’+b’), dp
The (not very interesting) figure at left shows that the replaced value of variable 5 agrees with the original value. This can be done for multiple variables.
In the second installment of this Missing Data series I’ll give some examples of how this works in practice, discuss limitations, and show some alternate ways of estimating missing values. In the third installment we’ll get to the much more challenging issue of building models on incomplete data sets.
BMW
B.M. Wise and N.L. Ricker, “Recent advances in Multivariate Statistical Process Control, Improving Robustness and Sensitivity,” IFAC Symposium n Advanced Control of Chemical Processes, pps. 125-130, Toulouse, France, October 1991.
Report from FACSS
Oct 6, 2011
This year’s Federation of Analytical Chemistry and Spectroscopy Societies meeting (FACSS) was quite vibrant. The number of participants was up from recent years, with close to 1200 registrants. Attendance at the technical sessions and traffic at the exhibit was good.
As usual, EVRI was there in force. Our booth crew is shown above, including me, Chief of Technology Development Jeremy M. Shaver, Vice-President Neal B. Gallagher, and Senior Research Scientist Randy Bishop.
 We especially enjoyed the Monday evening reception, where we gave away beer in our Eigenvector logo bottle koozies. Anna Cavinato of Eastern Oregon University is shown at left with a beer and koozie along with all the usual trade show accoutrements (including badge, necklace, wine glass, frisbee, product literature, t-shirt, etc.). We also made sure we were first in line on Tuesday when HORIBA Scientific gave away free hot dogs and beer for lunch.
We especially enjoyed the Monday evening reception, where we gave away beer in our Eigenvector logo bottle koozies. Anna Cavinato of Eastern Oregon University is shown at left with a beer and koozie along with all the usual trade show accoutrements (including badge, necklace, wine glass, frisbee, product literature, t-shirt, etc.). We also made sure we were first in line on Tuesday when HORIBA Scientific gave away free hot dogs and beer for lunch.
The organizers of FACSS also announced that the conference name has been changed to SCIX, short for Scientific Exchange. SCIX 2012 will be in Kansas City, Missouri.
As noted in the previous post, the Eigenvectorians also presented four talks, Jeremy co-taught the Analytical Raman Spectroscopy Workshop, Neal co-chaired a session on General Forensics, and Eigenvector sponsored sessions on Chemometrics and Data Fusion and Chemometrics for Process Analysis. On top of that there was booth duty, attending talks, reviewing posters, and the Raman and SAS receptions. It was a busy time!
EVRI would like to thank the organizers of FACSS 2011, especially Exhibits Chair Mike Carrabba, Workshop Chairs Brandye Smith-Goettler and Heather Brooke, and of course Cindi Lilly and her crew. Great meeting! See you next year!
BMW
Report from SIMS XVIII
Sep 26, 2011
The eighteenth meeting on Secondary Ion Mass Spectrometry, SIMS XVIII, was held last week in Riva del Garda, Italy. The meeting has been held biannually starting in 1977. As Nicolas Winograd pointed out in the opening lecture, it is a testament to the importance and vibrancy of the field that such a specialized meeting has continued to thrive. SIMS XVIII attracted nearly 400 participants divided roughly equally between Asia, the Americas, and Europe, with Asia being the somewhat largest third and the Americas the smallest.
I was pleased to see an increase in the number of papers that utilized multivariate analysis (MVA) in general and PLS_Toolbox and MIA_Toolbox in particular. In her talk, “Critical Issues in Multivariate Analysis of ToF SIMS Spectra, Images and Depth Profiles,” Bonnie Tyler noted that the number of publications in SIMS utilizing MVA is currently exponentiating, a trend which started about 10 years ago. Interestingly, I first taught a course utilizing SIMS data in 2001 at the Sanibel Island ASMS Meeting. That said, I still saw plenty of presentations and posters with 8-12 images at different AMUs that all looked the same: instances where PCA would result in a drastic reduction in the number of images to review plus some corresponding noise reduction.
Interesting talks where our software was used included:
- “ToF-SIMS Technique for Nano-Surface Analysis of Biosensors and Tissues” by Tae Geol Lee, Ji-Won Park, Sojeong Yun, Heesang Song, Hyegeun Min, Hyun Jo Jung, Taek Dong Chung, Ki-Chul Hwang, Hark Kyun Kim, Dae Won Moon and Daehee Hwang
- “Development of ToF-SIMS Enzyme Screening Assays” by Robyn Goacher, Elizabeth Edwards, Charles Mims and Emma Master
- “Evaluation of white radish sprouts growth influenced by magnetic fields using TOF-SIMS and MCR” by Satoka Aoyagi, Katsushi Kuroda, Ruka Takama, Kazuhiko Fukushima, Isao Kayano, Seiichi Mochizuki and Akira Yano
- “Multivariate Image Analysis of Chemical Heterogeneities Observed in Microarray Printed Polymers” by David Scurr, Andrew Hook, Daniel Anderson, Robert Langer, Morgan Alexander and Martyn Davies
I presented “Deconvolving SIMS Images using Multivariate Curve Resolution with Contrast Constraints,” by myself and Willem Windig. The talk demonstrated how in MCR contrast can be maximized in either the estimated spectra or concentrations/images. The solutions for each of these cases will give an indication of the range of solutions in the given MCR problem which all fit the data equally well. In many instances, one of the two solutions will be preferred, or both may be interpretable to illustrate different features of the data.
SIMS XVIII also included another installment of our “Chemometrics in SIMS” course. I was happy to present this course to another eager group of future chemometricians!
The SIMS XVIII conference was a great success. The technical program was very interesting and went off without a hitch. Riva del Garda is a beautiful location, perhaps the most scenic conference venue I have experienced. Congratulations to the organizers! I’m already looking forward to SIMS XIX in Jeju Island, Korea in 2013.
BMW
I hope this isn’t a trend
Jul 11, 2011
On April 21, I received an announcement and call for papers from the organizers of ICRM 2011. The announcement noted that the scientific committee would start to select the abstracts for oral and poster presentations starting May 1. On April 28 I submitted an abstract for an oral presentation and received an automated reply noting that a decision would be made by July 1.
On May 9 I received a note from the conference chair, which stated, “Only abstracts of those who have registered by May 20th and paid their fee within 30 days of registration (and at least before June 20th) will be considered by the scientific committee for acceptance.” When I asked the organizers what would happen if I registered for the conference, and then decided to withdraw if my paper was not accepted, the reply included the cancellation policy, which states: “Registrations cancelled after July 1st 2011 or for no-shows at the conference will remain payable at full charges.”
So, at ICRM, in order to have your paper considered, you must pay the conference registration in full. And if they decide not to accept it, and you decide not to attend, you won’t get a refund.
It seems to the me that the decision to accept a paper should be based on scientific merit, not financial considerations. At ICRM, the scientific committee is actually more of an economic committee. I sincerely hope this isn’t the start of a trend among scientific conferences.
BMW
Rasmus Bro Wins Wold Medal
Jun 9, 2011
The 10th Wold Medal was presented at the Scandinavian Symposium on Chemometrics, SSC12, in Billund, Denmark, this evening. Professor Rasmus Bro accepted the award from past winner Johan Trygg of Umeå University. Rasmus credited Lars Munck for development of the group at University of Copenhagen and enabling him to flourish there. He also mentioned the late Sijmen de Jong as being one of the great people he worked with and dedicated the award to him.
The Wold Medal, named for Herman Wold, father of Partial Least Squares (PLS) methods, honors individuals who have contributed greatly to the field of chemometrics. Rasmus is a worthy recipient, having published over 100 papers in the field, particularly in multi-way analysis. Prof. Bro is also know for his teaching, MATLAB-based multi-way software, and as being a great ambassador for the field. Rasmus is shown below with the Wold medal.
Past recipients of the Wold medal include Svante Wold, Agnar Höskuldsson, Harald Martens, John MacGregor, Rolf Carlson, Olav Kvalheim, Pentti Minkinen, Michael Sjöström and Johan Trygg.
Congratulations Rasmus! Well deserved!
BMW
EigenU Poster Session Winners
May 25, 2011
The Sixth Annual Eigenvector University was held last week, and with it, our Tuesday evening PLS_Toolbox User Poster Session. As always, the poster session allowed EigenU participants to discuss chemometric applications over hors d’oeuvres and drinks.
This year’s winners were Kellen Sorauf of University of Denver and Christoph Lenth of Laser-Laboratorium Göttingen e.V. Kellen, shown below receiving his iPod nano, presented Distribution Coefficient of Pharmaceuticls on Clay Mineral Nanoparticles Using Multivariate Curve Resolution with co-authors Keith E. Miller and Todd A. Wells.

Christoph, shown below, presented Qualitative trace analytics of proteins using SERS with co-authors W. Huttner, K. Christou and H. Wackerbarth.

Poster session winners received the traditional iPod nano, (16G), engraved with EigenU 2011 Best Poster.
Thanks to everyone who attended and presented!
BMW
Eigenvectorians Hit the Road
May 6, 2011
The next six months promise to be a busy time for the Eigenvectorians. We’ll be hitting the road to eight conferences in the US and Europe. I’ve listed below the conferences we’ll be attending, the talks and courses we’ll be giving, and our exhibits. (Note: not all talks have been accepted yet!)
SSC12, Billund, Denmark, June 7-10
- A Guide to Orthogonalization Filters Used in Multivariate Image Analysis and Conventional Applications, by Barry M. Wise (BMW), Jeremy M. Shaver (JMS) and Neal B. Gallagher (NBG)
- We will also offer Advanced Preprocessing at SSC12 on June 7
AOAC PacNW, Tacoma, Washington, June 21-22
- Simultaneous Analysis of Multivariate Images from Multiple Analytical Techniques, by JMS, and Eunah Lee of HORIBA Scientific
- We’ll also have an exhibit table at AOAC, so please stop by!
NORM 2011, Portland, Oregon June 26-29
- A Guide to the Orthogonalization Filter Smörgåsbord , by BMW, JMS and NBG
SIMS XVIII, Riva del Garda, Italy, September 18-23
- Deconvolving SIMS Images using Multivariate Curve Resolution with Contrast Constraints , by BMW and Willem Windig (WW)
ICRM, Nijmegen, The Netherlands, September 25-29
- Analysis of Hyperspectral Images using Multivariate Curve Resolution with Contrast Constraints , by BMW and WW
FACSS, Reno, Nevada, October 2-7
- Multivariate Modeling of Batch Processes Using Summary Variables, by BMW and NBG
- Detection of Adulterants in Raw Materials Utilizing Hyperspectral Imaging with Target and Anomaly Detection Algorithms, by Lam K. Nguyen and Eli Margilith of Opotek, NBG and JMS
- A Comparison of Detection Strategies for Solids and Organic Liquids on Surfaces Using Long Wave Infrared Hyperspectral Imaging, by NBG, and Thomas A. Blake and James F. Kelly of PNNL
- Visualizing Results: Data Fusion and Analysis for Hyperspectral Images, by JMS, Eunah Lee and Karen Gall
- EVRI will also offer Advanced Preprocessing and Intro to Multivariate Image Analysis at FACSS
- EVRI will also be at the FACSS Exhibit in Booth #29
AIChE Annual Meeting, Minneapolis, MN, October 16-21
- Correlating Powder Flowability to Particle Size Distribution using Chemometric Techniques by Christopher Burcham of Eli Lilly and Robert T. Roginski (RTR)
EAS, Somerset, NJ, November 14-17
- Orthogonalization Filter Preprocessing in NIR Spectroscopy, by BMW, JMS and NBG
- Eigenvector is once again proud to sponsor the EAS Award for Achievements in Chemometrics, this year honoring Beata Walczak
- BMW will assist Don Dahlberg with a new short course, Intermediate Chemometrics without Equations
- EVRI can also be found at the EAS Exposition in Booth #508
Now that you know where we’re going to be, we hope you’ll stop by and say hello!
BMW
iPods Ordered!
Apr 25, 2011
EigenU 2011 is now less than 3 weeks away: it starts on Sunday, May 15. Once again we’ll have the “PLS_Toolbox/Solo User Poster Session” where users can showcase their own chemometric achievements. This Tuesday evening event is open to all EigenU participants, as well as anyone with a poster which demonstrates how PLS_Toolbox or Solo was used in their work. Presenters of the two best posters will receive Apple iPod nanos for their effort. I ordered them today, (16G, one blue, one orange), engraved with EigenU 2011 Best Poster.
Tuesday evening will also include a short User Group meeting. We’ll give a short presentation highlighting the key features in our latest software releases and discuss directions for future development. Here is your chance to give us your wish list!
The poster session will feature complimentary hors d’oeuvres and beverages. This is a great time to mingle with colleagues and the Eigenvectorians and discuss the many ways in which chemometric methods are used.
See you in May!
BMW
Switch from MLR to PLS?
Apr 20, 2011
In a recent post, Fernando Morgado posed the following question to the NIR Discussion List: “When is it necessary to move from traditional Multiple Linear Regression (MLR) to Partial Least Squares Regression (PLS)?” That’s a good discussion starter! I’ll offer my take on it here. I have several answers to the question, but before I get to that, it is useful to outline the differences between MLR and PLS.
MLR, PLS, Principal Components Regression (PCR) and a number of other methods are all Inverse Least Squares (ILS) models. Given a set of predictors, X (m samples by n variables), and a variable to be predicted, y (m by 1), they find b, such that the estimate of y, ý = Xb, from b = X+y. The difference between the methods is that they all use different ways to estimate X+, the pseudoinverse of X.
In MLR, X+ = (XTX)-1XT. In PCR, where the data matrix is decomposed via Principal Components Analysis (PCA) as X = TkPkT + E, where Tk is the (m by k) matrix containing the scores on the first k PCs, Pk is the (n by k) matrix containing the first k PC loadings and E is a matrix of residuals, then X+ = Pk(TkTTk)-1TkT. The number of PCs, k, is determined via cross-validation or any number of other methods. In PLS, the decomposition of X is somewhat more complicated, and the resulting inverse is X+ = Wk(PkTWk)-1(TkTTk)-1TkT, where the additional parameter Wk (n by k) is known as the weights.
With that background covered, we can now consider “When is it necessary to move from traditional Multiple Linear Regression (MLR) to Partial Least Squares Regression (PLS)?”
1) Any time the rank of the spectral data matrix X is less than the number of variables. The mathematical rank of a matrix is well defined as the number of linearly independent rows or columns. It is important because the MLR solution includes the term (XTX)-1 in the pseudoinverse. If X has rank less than the number of variables, XTX has rank less than its dimension, i.e. is rank deficient, and its inverse is undefined. PCR and PLS avoid this problem by decomposing X and forming a solution based on the large variance (stable) part of the decomposition. From this, it is clear that another answer to the question must be:
2) Any time the data contains fewer samples than variables. This is a common problem in spectroscopy because many instruments measure hundreds or thousands of variables (channels), but acquiring that many samples can be a very expensive proposition. The obvious follow on question is, “Then why not just reduce the number of variables.” The answer to that is, in short, noise reduction. Averaging of correlated measurements results in a reduction in noise.
But what about the case with m > n, but X is nearly rank deficient? That is, X is of full rank only because it is corrupted by noise? This leads to:
3) Any time the chemical rank of the spectral data matrix X is less than the number of variables. By chemical rank we mean the number of variations in the data that are due to chemical variation as opposed to detector noise and other minor effects.
So if any of the above three conditions exist, then it is appropriate to move from MLR to a factor-based method such as PLS or PCR. But I’m going to play devil’s advocate here a bit and give one more answer:
4) Always. MLR is, after all, just a special case of PLS and PCR. If you include all the components, you arrive at the same model that MLR gives you. But along the way you get additional diagnostic information from which you might learn something. The scores and loadings (and weights) all give you information about the chemistry.
On the NIR Discussion list, Donald J. Dahm wrote: “As a grumpy old man, I say the time to switch to PLS is when you are ready to admit that you don’t have the knowledge or patience to do actual spectroscopy.” I hope that was said with tongue firmly planted in cheek, because I’d argue that the opposite is true. When you are using PLS or PCR and interrogating the models, you are learning about the chemistry. When you use MLR, you are simply fitting the data.
BMW
“This software is awesome!”
Apr 7, 2011
That’s what Eigenvector’s Vice-President, Neal B. Gallagher, wrote to our developers, Jeremy, Scott and Donal, this morning about the latest version of PLS_Toolbox. So, OK, he may be a little biased. But it underscores a point, which is that all of us at EVRI who do consulting (including Neal, Jeremy, Bob, Randy, Willem and myself) use our software constantly to do the same type of tasks our users must accomplish. We write it for our own use as well as theirs. (Scott pointed out that this is known as “eating your own dog food.”)
Neal went on to say, “Every time I turn around, this package and set of solutions just keeps getting better and more extensive. I am truly convinced that this is the most useful package of its kind on the market. This is due to your intelligence, foresight, and attention to detail. It is also due to your open mindedness and willingness to listen to your users.”
Thanks, Neal. I think you made EVRIbody’s day!
BMW
Improved Model_Exporter Coming Soon
Mar 29, 2011
EVRI’s Model_Exporter allows users of PLS_Toolbox and Solo to export their Principal Component Analysis (PCA), Partial Least Squares (PLS) Regression and other models in a variety of formats suitable for use in both MATLAB and other environments (e.g. LabSpec, GNU Octave, Symbion, LabVIEW, Java, C++/C#). A new, improved version of Model_Exporter will be released in the coming weeks. The new version will allow models to be more easily applied on platforms with less computer power, such as on hand-held devices.
The new Model_Exporter will include a number of improvements including a much less memory-intensive encoding of Savitzky-Golay derivatives and smoothing and similar improvements in handling of excluded variables. The new version will also support additional preprocessing methods which were released in versions 6.0 and 6.2 of PLS_Toolbox and Solo.
The other big news is that we will also be bundling with Model_Exporter a freely-distributable .NET class to do predictions from exported models. As-is, this class can be integrated into any .NET application (C++/C#/VB) from Microsoft Visual Studio 2005 or later. It can be used and redistributed without any per-use charge (exactly as the exported models are licensed) and we’d be happy to work with you on integrating this class into your end application. If you have interest in this approach, just let our Chief of Technology Development, Jeremy Shaver, or me know.
BMW
PLS_Toolbox or Solo?
Mar 28, 2011
Customers who are new to chemometrics often ask us if they should purchase our MATLAB-based PLS_Toolbox or stand-alone Solo. I’ll get to some suggestions in a bit, but first it is useful to clarify the relationship between the two packages: Solo is the compiled version of the PLS_Toolbox tools which are accessible from the interfaces. Similarly, Solo+MIA is the compiled version of PLS_Toolbox and MIA_Toolbox. The packages share the same code-base, which is great for the overall stability of the software as they operate identically. They are also completely compatible, so PLS_Toolbox and Solo users can share data, models, preprocessing, and results. Models from each are equally easily applied on-line using Solo_Predictor.
The majority of the functionality of PLS_Toolbox is accessible in Solo, particularly the most commonly used tools. Every method that you access through the main Graphical User Interfaces (GUIs) of PLS_Toolbox is identical in Solo. But with Solo you don’t get the MATLAB command line, so there are some command-line only functions that are not accessible. The number of these, however, is decreasing all the time as we make more PLS_Toolbox functionality accessible through the GUIs. And of course all the GUIs in PLS_Toolbox + MIA_Toolbox are accessible in Solo+MIA.
So what to buy? Well, do you already have access to MATLAB? Over one million people do, and it is available in many universities and large corporations. If you have access to MATLAB, then buy PLS_Toolbox (and MIA_Toolbox). It costs less than Solo (and Solo+MIA), includes all the functionality of Solo and then some, can be accessed via command line and called in MATLAB scripts and functions.
And if you don’t already have MATLAB? If you only need to use the mainstream modeling and analysis functions, then Solo (and Solo+MIA) will save you some money over purchasing MATLAB and PLS_Toolbox (and MIA_Toolbox). I’d only purchase MATLAB if I needed to write custom scripts and functions that call PLS_Toolbox functions. Honestly, the vast majority of our users can do what they need to do using the GUIs. The stuff you can’t get to from the GUIs is pretty much just for power users.
That said, the big plus about buying MATLAB plus PLS_Toolbox is that you get MATLAB, which is a tremendously useful tool in its own right. Once you start using it, you’ll find lots of things to do with it besides just chemometrics.
Hope that helps!
BMW
EigenU 2011 Registration Now Open
Jan 25, 2011
The Sixth edition of Eigenvector University will be held May 15-20, 2011. Once again, we’ll be at the fabulous Washington Athletic Club in the heart of downtown Seattle.
We’ve added two new classes this year, “SVMs, LWR and other Non-linear Methods for Calibration and Classification” and “Design of Experiments.” The non-linear methods class will focus on Support Vector Machines for regression and classification, which were added to PLS_Toolbox/Solo version 5.8 (February 2010) along with an interface to Locally Weighted Regression. We’ve found these methods to be quite useful in a number of situations, as have our users. The DOE course will focus on practical aspects experimental design, including designing data sets for multivariate calibration.
I sometimes say that the secret to Eigenvector Research is data preprocessing, i.e. what you do to the data before it hits a PLS or other multivariate model. Thus, “Advanced Preprocessing” has been expanded to a full day for EigenU 2011. We’ll cover many methods for eliminating extraneous variance, including the “decluttering” methods (Generalized Least Squares Weighting, External Parameter Orthogonalization, etc.) we’ve highlighted recently. “Multivariate Curve Resolution” has also be expanded to a full day in order to better cover the use of constraints and contrast control.
EigenU 2011 will also include three evening events, including Tuesday night’s “PLS_Toolbox/Solo User Poster Session,” (with iPod Nanos for the best two posters), Wednesday night’s “PLS_Toolbox/Solo PowerUser Tips & Tricks,” and the Thursday evening dinner event.
You can register for EigenU through your user account. For early discount registration, payment must be received by April 15. Questions? E-mail me.
See you at EigenU!
BMW
Sijmen de Jong
Nov 8, 2010
Sijmen de Jong passed away on October 30, 2010.
I first heard of Sijmen not long after finishing my dissertation. It was late 1992 or so, and I was working at Battelle, Pacific Northwest National Laboratory (PNNL). Sijmen had gotten a copy of an early version of PLS_Toolbox which was available on the internet via FTP. I received a letter from Sijmen with a number of “suggestions” as to how the toolbox might be improved. As I recall, the letter ran at least 3 pages, and included two floppy disks (5.25 inch!) filled with MATLAB .m files.
I was initially taken aback by the letter, and recall thinking, “Who is this guy?” But it didn’t take me long to figure out that, wow!, there was a lot of good stuff there. Several of the routines were incorporated into the next release of PLS_Toolbox, and modifications were made to several others. I’m sure there is still code of his in the toolbox today!
Sijmen was especially interested in the work I’d done with Larry Ricker on Continuum Regression (CR) [1], as he had just published on another continuously adjustable technique, Principal Covariates Regression (PCovR) [2]. He was also working on his SIMPLS [3] algorithm for PLS around that time. Sijmen suggested that the SIMPLS algorithm might be extended to CR. This started a collaboration which would eventually produce the paper “Canonical partial least squares and continuum power regression” in 2001 [4]. I still think that this is the best paper I’ve ever had the pleasure of being associated with. It won the Unilever R&D Vlaardingen “Author Award,” and I still display the certificate proudly on my office wall.
While CR is primarily of academic interest, SIMPLS has become perhaps the most widely used PLS regression algorithm. The reasons for this are evident from several of my recent posts concerning accuracy and speed of various algorithms. If you have done a PLS regression in PLS_Toolbox or Solo, you have benefited from Sijmen’s work!
Sijmen was the kind of smart that I always wanted to be. He seemed to see clearly through the complex math, understand how methods are related, and see how a small “trick” might greatly simplify a problem. As another colleague put it, “A very clever guy!” Beyond that, he was easy to work with and fun to be around. I regret that I got to be around him socially only a few times.
Rest in peace, Sijmen. You will be missed!
BMW
[1] B.M. Wise and N.L. Ricker, “Identification of Finite Impulse Response Models with Continuum Regression,” J. Chemometrics, 7(1), pps. 1-14, 1993.
[2] S. de Jong, H.A.L. Kiers, “Principal Covariates Regression: Part 1, Theory,” Chemo. and Intell. Lab. Sys., Vol 14, pps. 155-164, 1992.
[3] S. de Jong, “SIMPLS: an alternative approach to partial least squares regression,” Chemo. and Intell. Lab. Sys., Vol. 18, pps. 251-263, 1993.
[4] S. de Jong, B.M. Wise and N.L. Ricker, “Canonical partial least squares and continuum power regression,” J. Chemometrics, 15(1), pps. 85-100, 2001.
Speed of PLS Algorithms
Nov 1, 2010
Previously I wrote about accuracy of PLS algorithms, and compared SIMPLS, NIPALS, BIDIAG2 and the new DSPLS. I now turn to speed of the algorithms. In the paragraphs that follow I’ll compare SIMPLS, NIPALS and DSPLS as implemented in PLS_Toolbox 6.0. It should be noted that the code I’ll test is our standard code (PLS_Toolbox functions simpls.m, dspls.m and nippls.m). These are not stripped down algorithms. They include all the error trapping (dimension checks, ranks checks, etc.) required to use these algorithms with real data. I didn’t include BIDIAG2 here because we don’t support it, and as such, I don’t have production code for it, just the research code (provided by Infometrix) I used to investigate BIDIAG2 accuracy. The SIMPLS and DSPLS code used here includes the re-orthoginalization step investigated previously.
The tests were performed on my (4 year old!) MacBook Pro laptop, with a 2.16GHz Intel Core Duo, and 2GB RAM running MATLAB 2009b (32 bit). The first figure, below, shows straight computation time as a function of number of samples for SIMPLS to calculate a 20 Latent Variable (LV) model, for data with 10 to 1,000,000 variables (legend gives number of variables for each line). The maximum size of X for each run was 30 million elements, so the lines all terminate at this point. Times range from a minimum of ~0.003 seconds to just over 10 seconds.

It is interesting to note that, for the largest X matrices the times vary from 4 to 12 seconds, with the faster times being for the more square X‘s. It is fairly impressive, (at least to me), that problems of this size are feasible on an outdated laptop! A 10-way split cross-validation could be done in less than a minute for most of the large cases.
The second figure shows the ratio of the computation time of NIPALS to SIMPLS as a function of number of samples. Each line is a fixed number of variables (indicated by the legend). Maximum size of X here is 9 million elements (I just didn’t want to wait for NIPALS on the big cases). Note that SIMPLS is always faster than NIPALS. The difference is relatively small (around a factor of 2) for the tall skinny cases (many samples, few variables) but considerable for the short fat cases (few samples, many variables). For the case of 100 samples and 100,000 variables, SIMPLS is faster than NIPALS by more than a factor of 10.

The ratio of computation time for DSPLS to SIMPLS is shown in the third figure. These two methods are quite comparable, with the difference always less than a factor of 2. Thus I’ve chosen to display the results as a map. Note that each method has its sweet spot. SIMPLS is faster (red squares) for the many variable problems while DSPLS is faster (light blue squares) for the many sample problems. (Dark blue area represents models not computed for X larger than 30 million elements). Overall, SIMPLS retains a slight time advantage over DSPLS, but for the most part they are equivalent.

Given the results of these tests, and the previous work on accuracy, it is easy to see why SIMPLS remains our default algorithm, and why we found it useful to include DSPLS in our latest releases.
BMW

 SEARCH
SEARCH
