Category Archives: Software
New Versions Released
Mar 23, 2011
New versions of our most popular software products were released earlier this month. Our MATLAB-based, full-featured chemometrics package, PLS_Toolbox, is now in version 6.2. The stand-alone version of PLS_Toolbox, Solo, and Solo+MIA for multivariate and hyperspectral image analysis also move to version 6.2. MIA_Toolbox is now in version 2.5.
The functionality of Solo+MIA and MIA_Toolbox has been expanded greatly with the integration of ImageJ, the image processing application developed by NIH. New tools include particle counting with size and shape analysis, interactive data navigator for drilling into an image, magnify, cross-section and image alignment. A number of new data importers have also been added (RAW, BIF and ENVI).
For a tour of the new features, highlighting the new MIA tools, please watch this short video. For a complete list of new features, please see the PLS_Toolbox/Solo 6.2 release notes, and the MIA_Toolbox/Solo+MIA release notes.
Users with current maintenance contracts can download the new versions from their account. Free 30-day demos are also available for download.
We trust that the new tools will make you even more productive!
BMW
Speed of PLS Algorithms
Nov 1, 2010
Previously I wrote about accuracy of PLS algorithms, and compared SIMPLS, NIPALS, BIDIAG2 and the new DSPLS. I now turn to speed of the algorithms. In the paragraphs that follow I’ll compare SIMPLS, NIPALS and DSPLS as implemented in PLS_Toolbox 6.0. It should be noted that the code I’ll test is our standard code (PLS_Toolbox functions simpls.m, dspls.m and nippls.m). These are not stripped down algorithms. They include all the error trapping (dimension checks, ranks checks, etc.) required to use these algorithms with real data. I didn’t include BIDIAG2 here because we don’t support it, and as such, I don’t have production code for it, just the research code (provided by Infometrix) I used to investigate BIDIAG2 accuracy. The SIMPLS and DSPLS code used here includes the re-orthoginalization step investigated previously.
The tests were performed on my (4 year old!) MacBook Pro laptop, with a 2.16GHz Intel Core Duo, and 2GB RAM running MATLAB 2009b (32 bit). The first figure, below, shows straight computation time as a function of number of samples for SIMPLS to calculate a 20 Latent Variable (LV) model, for data with 10 to 1,000,000 variables (legend gives number of variables for each line). The maximum size of X for each run was 30 million elements, so the lines all terminate at this point. Times range from a minimum of ~0.003 seconds to just over 10 seconds.

It is interesting to note that, for the largest X matrices the times vary from 4 to 12 seconds, with the faster times being for the more square X‘s. It is fairly impressive, (at least to me), that problems of this size are feasible on an outdated laptop! A 10-way split cross-validation could be done in less than a minute for most of the large cases.
The second figure shows the ratio of the computation time of NIPALS to SIMPLS as a function of number of samples. Each line is a fixed number of variables (indicated by the legend). Maximum size of X here is 9 million elements (I just didn’t want to wait for NIPALS on the big cases). Note that SIMPLS is always faster than NIPALS. The difference is relatively small (around a factor of 2) for the tall skinny cases (many samples, few variables) but considerable for the short fat cases (few samples, many variables). For the case of 100 samples and 100,000 variables, SIMPLS is faster than NIPALS by more than a factor of 10.

The ratio of computation time for DSPLS to SIMPLS is shown in the third figure. These two methods are quite comparable, with the difference always less than a factor of 2. Thus I’ve chosen to display the results as a map. Note that each method has its sweet spot. SIMPLS is faster (red squares) for the many variable problems while DSPLS is faster (light blue squares) for the many sample problems. (Dark blue area represents models not computed for X larger than 30 million elements). Overall, SIMPLS retains a slight time advantage over DSPLS, but for the most part they are equivalent.

Given the results of these tests, and the previous work on accuracy, it is easy to see why SIMPLS remains our default algorithm, and why we found it useful to include DSPLS in our latest releases.
BMW
One Last Time on Accuracy of PLS Algorithms
Sep 9, 2010
Scott Ramos of Infometrix wrote me last week and noted that he had followed the discussion on the accuracy of PLS algorithms. Given that they had done considerable work comparing their BIDIAG2 implementation with other PLS algorithms, he was “surprised” with my original post on the topic. He was kind enough to send me a MATLAB implementation of the PLS algorithms included in their product Pirouette for inclusion in my comparison.
Below you’ll find a figure that compares regression vectors from NIPALS, SIMPLS, DSPLS and the BIDIAG2 code provided by Scott. The figure was generated for a tall-skinny X-block (our “melter” data) and for a short-fat X-block (NIR of pseudo-gasoline mixture). Note that I used our SIMPLS and DSPLS that include re-orthogonalization, as that is now our default. While I haven’t totally grokked Scott’s code, I do see where it includes an explicit re-orthogonalization of the scores as well.

Note that all the algorithms are quite similar, with the biggest differences being less than one part in 1010. The BIDIAG2 code provided by Scott (hot pink with stars) is the closest to NIPALS for the tall skinny case, while being just a little more different than the other algorithms for the short fat case.
This has been an interesting exercise. I’m always surprised when I find there is still much to learn about something I’ve already been thinking about for 20+ years! It is certainly a good lesson in watching out for numerical accuracy issues, and in how accuracy can be improved with some rather simple modifications.
BMW
Coming in 6.0: Analysis Report Writer
Sep 4, 2010
Version 6.0 of PLS_Toolbox, Solo and Solo+MIA will be out this fall. There will be lots of new features, but right at the moment I’m having fun playing with one in particular: the Analysis Report Writer (ARW). The ARW makes documenting model development easy. Once you have a model developed, just make the plots you want to make, then select Tools/Report Writer in the Analysis window. From there you can select HTML, or if you are on a Windows PC, MS Word or MS Powerpoint for saving your report. The ARW then writes a document with all the details of your model, along with copies of all the figures you have open.
As an example, I’ve developed a (quick and dirty!) model using the tablet data from the IDRC shootout in 2002. The report shows the model details, including the preprocessing and wavelength selection, along with the plots I had up. This included the calibration curve, cross-validation plot, scores on first two PCs, and regression vector. But it could include any number and type of plots as desired by the user. Statistics on the prediction set are also included.
Look for Version 6.0 on our web site in October, or come visit us at FACSS, booth 48.
BMW
Advanced Features in Barcelona
Sep 1, 2010
I’m pleased to announce that I’ll be doing a one day short course, Using the Advanced Features of PLS_Toolbox, on the University of Barcelona campus, on October 1, 2010 (one month from today!). The course will show how to use many of the powerful, but often underutilized, tools in PLS_Toolbox. It will also feature some new tools from the upcoming PLS_Toolbox 6.0, to be released this fall.
Normally, our courses focus on developing an understanding of chemometric methods such as PCA and PLS. This course is something of a departure in that it focuses on getting the most out of the software. This will give us a chance to show how to access many of the advanced features and methods implemented in PLS_Toolbox.
You can get complete information, including registration info and a course outline, on the course description page. If you have additional questions or suggestions for demos you’d like to see, contact me.
See you in Barcelona!
BMW
Re-orthogonalization of PLS Algorithms
Aug 23, 2010
Thanks to everybody that responded to the last post on Accuracy of PLS Algorithms. As I expected, it sparked some nice discussion on the chemometrics listserv (ICS-L)!
As part of this discussion, I got a nice note from Klaas Faber reminding me of the short communication he wrote with Joan Ferré, “On the numerical stability of two widely used PLS algorithms.” [1] The article compares the accuracy of PLS via NIPALS and SIMPLS as measured by the degree to which the scores vectors are orthogonal. It finds that SIMPLS is not as accurate as NIPALS, and suggests adding a re-orthogonalization step to the algorithms.
I added re-orthogonalization to the code for SIMPLS, DSPLS, and Bidiag2 and ran the tests again. The results are shown below. Adding this to Bidiag2 produces the most dramatic effect (compare to figure in previous post), and improvement is seen with SIMPLS as well. Now all of the algorithms stay within ~1 part in 1012 of each other through all 20 LVs of the example problem. Reconstruction of X from the scores and loadings (or weights, as the case may be!) was improved as well. NIPALS, SIMPLS and DSPLS all reconstruct X to within 3e-16, while Biadiag2 comes in at 6e-13.
The simple fix of re-orthogonalization makes Bidiag2 behave acceptably. I certainly would not use this algorithm without it! Whether to add it to SIMPLS is a little more debatable. We ran some tests on typical regression problems and found that the difference on predictions with and without it was typically around 1 part in 108 for models with the correct number of LVs. In other words, if you round your predictions to 7 significant digits or less, you’d never see the difference!
That said, we ran a few tests to determine the effect on computation time of adding the re-orthogonalization step to SIMPLS. For the most part, the effect was negligible, less than 5% additional computation time for the vast majority of problem sizes and at worst a ~20% increase. Based on this, we plan to include re-orthogonalization in our SIMPLS code in the next major release of PLS_Toolbox and Solo, Version 6.0, which will be out this fall. We’ll put it in the option to turn it off, if desired.
BMW
[1] N.M. Faber and J. Ferré, “On the numerical stability of two widely used PLS algorithms,” J. Chemometrics, 22, pps 101-105, 2008.
Accuracy of PLS Algorithms
Aug 13, 2010
In 2009 Martin Andersson published “A comparison of nine PLS1 algorithms” in Journal of Chemometrics[1]. This was a very nice piece of work and of particular interest to me as I have worked on PLS algorithms myself [2,3] and we include two algorithms (NIPALS and SIMPLS) in PLS_Toolbox and Solo. Andersson compared regression vectors calculated via nine algorithms using 16 decimal digits (aka double precision) in MATLAB to “precise” regression vectors calculated using 1000 decimal digits. He found that several algorithms, including the popular Bidiag2 algorithm (developed by Golub and Kahan [4] and adapted to PLS by Manne [5]), deviated substantially from the vectors calculated in high precision. He also found that NIPALS was among the most stable of algorithms, and while somewhat less accurate, SIMPLS was very fast.
Andersson also developed a new PLS algorithm, Direct Scores PLS (DSPLS), which was designed to be accurate and fast. I coded it up, adapted it to multivariate Y, and it is now an option in PLS_Toolbox and Solo. In the process of doing this I repeated some of Andersson’s experiments, and looked at how the regression vectors calculated by NIPALS, SIMPLS, DSPLS and Bidiag2 varied.
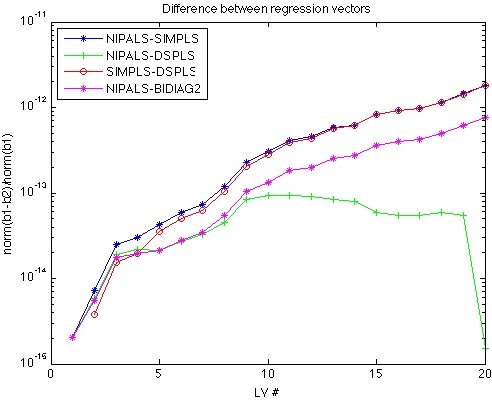
The figure below shows the difference between various regression vectors as a function of Latent Variable (LV) number for Melter data set, where X is 300 x 20. The plotted values are the norm of the difference divided by the norm the first regression vector. The lowest line on the plot (green with pluses) is the difference between the NIPALS and DSPLS regression vectors. These are the two methods that are in the best agreement. DSPLS and NIPALS stay within 1 part in 1012 out through the maximum number of LVs (20).

The next line up on the plot (red line and circles with blue stars) is actually two lines, the difference between SIMPLS and both NIPALS and DSPLS. These lie on top of each other because NIPALS and DSPLS are so similar to each other. SIMPLS stays within one part in 1010 through 10 LVs (maximum LVs of interest in this data) and degrades to one part in ~107.
The highest line on the plot (pink with stars) is the difference between NIPALS and Bidiag2. Note that by 9 LVs this difference has increased to 1 part in 100, which is to say that the regression vector calculated by Bidiag2 has no resemblance to the regression vector calculated by the other methods!
I programmed my version of Bidiag2 following the development in Bro and Eldén [6]. Perhaps there exist more accurate implementations of Bidiag2, but my results resemble those of Andersson quite closely. You can download my bidiag.m file, along with the code that generates this figure, check_PLS_reg_accuracy.m. This would allow you to reproduce this work in MATLAB with a copy of PLS_Toolbox (a demo would work). I’d be happy to incorporate an improved version of Bidiag in this analysis, so if you have one, send it to me.
BMW
[1] Martin Andersson, “A comparison of nine PLS1 algorithms,” J. Chemometrics, 23(10), pps 518-529, 2009.
[2] B.M. Wise and N.L. Ricker, “Identification of Finite Impulse Response Models with Continuum Regression,” J. Chemometrics, 7(1), pps 1-14, 1993.
[3] S. de Jong, B. M. Wise and N. L. Ricker, “Canonical Partial Least Squares and Continuum Power Regression,” J. Chemometrics, 15(2), pps 85-100, 2001.
[4] G.H. Golub and W. Kahan, “Calculating the singular values and pseudo-inverse of a matrix,” SIAM J. Numer. Anal., 2, pps 205-224, 1965.
[5] R. Manne, “Analysis of two Partial-Least-Squares algorithms for multivariate calibration,” Chemom. Intell. Lab. Syst., 2, pps 187–197, 1987.
[6] R. Bro and L. Eldén, “PLS Works,” J. Chemometrics, 23(1-2), pps 69-71, 2009.
Clustering in Images
Aug 6, 2010
It is probably an understatement to say that here are many methods for cluster analysis. However, most clustering methods don’t work well for large data sets. This is because they require computation of the matrix that defines the distance between all the samples. If you have n samples, then this matrix is n x n. That’s not a problem if n = 100, or even 1000. But in multivariate images, each pixel is a sample. So a 512 x 512 image would have a full distance matrix that is 262,144 x 262,144. This matrix would have 68 billion elements and take 524G of storage space in double precision. Obviously, that would be a problem on most computers!
In MIA_Toolbox and Solo+MIA there is a function, developed by our Jeremy Shaver, which works quite quickly on images (see cluster_img.m). The trick is that it chooses some unique seed points for the clusters by finding the points on the outside of the data set (see distslct_img.m), and then just projects the remaining data onto those points (normalized to unit length) to determine the distances. A robustness check is performed to eliminate outlier seed points that result in very small clusters. Seed points can then be replaced with the mean of the groups, and the process repeated. This generally converges quite quickly to a result very similar to knn clustering, which could not be done in a reasonable amount of time for large images.

 As an example, I’ve used the clustering function on a Landsat image of the Mississippi River. This 512 x 512 image has 7 channels. Working from the purely point-and-click Analysis interface on my 4 year old MacBook Pro laptop, this image can be clustered into 3 groups in 6.8 seconds. The result is shown at the far left. Clustering into 6 groups takes just a bit longer, 13.8 seconds. Results for the 6 cluster analysis are shown at the immediate left. This is actually a pretty good rendering of the different surface types in the image.
As an example, I’ve used the clustering function on a Landsat image of the Mississippi River. This 512 x 512 image has 7 channels. Working from the purely point-and-click Analysis interface on my 4 year old MacBook Pro laptop, this image can be clustered into 3 groups in 6.8 seconds. The result is shown at the far left. Clustering into 6 groups takes just a bit longer, 13.8 seconds. Results for the 6 cluster analysis are shown at the immediate left. This is actually a pretty good rendering of the different surface types in the image.
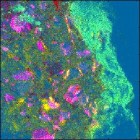
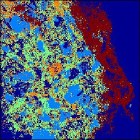 As another example, I’ve used the image clustering function on the SIMS image of a drug bead. This image is 256 x 256 and has 93 channels. For reference, the (contrast enhanced) PCA score image is shown at the far left. The drug bead coating is the bright green strip to the right of the image, while the active ingredient is hot pink. The same data clustered into 5 groups is shown to the immediate right. Computation time was 14.7 seconds. The same features are visible in the cluster image as the PCA, although the colors are swapped: the coating is dark brown and the active is bright blue.
As another example, I’ve used the image clustering function on the SIMS image of a drug bead. This image is 256 x 256 and has 93 channels. For reference, the (contrast enhanced) PCA score image is shown at the far left. The drug bead coating is the bright green strip to the right of the image, while the active ingredient is hot pink. The same data clustered into 5 groups is shown to the immediate right. Computation time was 14.7 seconds. The same features are visible in the cluster image as the PCA, although the colors are swapped: the coating is dark brown and the active is bright blue.
Thanks, Jeremy!
BMW
10,000 Commits
Jul 30, 2010
The EVRI software developers surpassed a landmark when our Donal O’Sullivan made the 10,000th “commit” to our software repository. Donal was working on some improvements to our Support Vector Machine (SVM) routines in PLS_Toolbox and submitted the changes yesterday afternoon.
As noted by our Chief of Technology Development, Jeremy Shaver, “This is a trivial landmark in some ways (it is just a number, like when your car rolls over 10,000 miles) but it also indicates just how active our development is. It all started with a revision by Scott in March, 2004. In six years time, we’ve committed thousands upon thousands of lines of code and many megabytes of files. That’s an average of 1666 commits/year or 4.6 commits/day although nearly 2000 of those were in the last year.”
The level of activity in our software repository truly demonstrates how our product development continues to accelerate. Thanks go to our users for driving, and funding, the advancements. And of course, to our developers, thanks for all your efforts, guys!
BMW
Robust Methods
May 12, 2010
This year we are presenting “Introduction to Robust Methods” at Eigenvector University. I’ve been working madly preparing a set of course notes. And I must say that it has been pretty interesting. I’ve had a chance to try the robust versions of PCA, PCR and PLS on many of the data sets we’ve used for teaching and demoing software, and I’ve been generally pleased with the results. Upon review of my course notes, our Donal O’Sullivan asked why we don’t use the robust versions of these methods all the time. I think that is a legitimate question!
In a nutshell, robust methods work by finding the subset of samples in the data that are most consistent. Typically this involves use of the Minimum Covariance Determinant (MCD) method, which finds the samples that have a covariance with the smallest determinant, which is a measure of the volume occupied by the data. The user specifies the fraction, h, to include, and the algorithm searches out the optimal set. The parameter h is between 0.5 and 1, and a good general default is 0.75. With h = 0.75 the model can resist up to 25% arbitrarily bad samples without going completely astray. After finding the h subset, the methods then look to see what remaining samples fall within the statistical bounds of the model and re-include them. Any remaining samples are considered outliers.
The main advantage of robust methods is that they automate the process of finding outliers. This is especially convenient when the data sets have many samples and a substantial fraction of bad data. How many times have you removed an obvious outlier from a data set only to find another outlier that wasn’t obvious until the first one is gone? This problem, known as masking, is virtually eliminated with robust methods. Swamping, when normal samples appear as outliers due to the confidence limits being stretched by the true outliers, is also mitigated.
So am I ready to set my default algorithm preferences to “robust?” Well, not quite. There is some chance that useful samples, sometimes required for building the model over a wide range of the data, will be thrown out. But I think I’ll at least review the robust results now each time I make a model on any medium or large data set, and consider why the robust method identifies them as outliers.
FYI, for those of you using PLS_Toolbox or Solo, you can access the robust option in PCA, PCR and PLS from the analysis window by choosing Edit/Options/Method Options.
Finally, I should note that the robust methods in our products are there due to a collaboration with Mia Hubert and her Robust Statistics Group at Katholieke Universiteit Leuven, and in particular, Sabine Verboven. They have been involved with the development of LIBRA, A MATLAB LIBrary for Robust Analysis. Our products rely on LIBRA for the robust “engines.” Sabine spent considerable time with us helping us integrate LIBRA into our software. Many thanks for that!
BMW
Welcome to the 64-bit party!
May 3, 2010
Unscrambler X is out, and CAMO is touting the fact that it is 64-bit. We say, “Welcome to the party!” MATLAB has had 64-bit versions out since April, 2006. That means that users of our PLS_Toolbox and MIA_Toolbox software have enjoyed the ability to work with data sets larger than 2Gb for over 4 years now. Our stand-alone packages, Solo and Solo+MIA have been 64-bit since June, 2009.
And how much is Unscrambler X? They don’t post their prices like we do. So go ahead and get a quote on Unscrambler, and then compare. We think you’ll find our chemometric software solutions to be a much better value!
BMW
Chemometric “how to” videos on-line
Nov 12, 2009
If a picture is worth a thousand words, what’s a video worth?
Here at EVRI we’ve started developing a series of short videos that show “how to” do various chemometric tasks with our software packages, including our new PLS_Toolbox 5.5 and Solo 5.5. Some of the presentations are pretty short and specific, but others are a little longer (10-15 minutes) and are a blend of teaching a bit about the method being used while showing how to do it in the software.
An example of the latter is “PCA on Wine Data” which shows how to build a Principal Components Analysis model on a small data set concerning the drinking habits, health and longevity of the population of 10 countries. Another movie, “PLS on Tablet Data” demonstrates building a Partial Least Squares calibration for a NIR spectrometer to predict assay values in pharmaceutical tablets.
While I was at it, I just couldn’t help producing a narrated version of our “Eigenvector Company Profile.”
We plan to have many more of these instructional videos that cover aspects of chemometrics and our software from basic to advanced. We hope you find each of them worth at least 1000 words!
BMW
The Gang’s all here: Chemometric software updates released
Nov 5, 2009
In this case, “the Gang” is all our most popular software packages, and they’ve all gotten substantial improvements. This includes our flagship MATLAB toolbox for chemometrics, PLS_Toolbox, and its stand-alone version Solo. Plus our products for Multivariate Image Analysis MIA_Toolbox and the stand alone Solo+MIA.
As evidenced by the release notes, PLS_Toolbox and Solo received a host of additions and improvements. I’m particularly geeked about the performance improvements we’ve made to our Multivariate Curve Resolution (MCR) code, which has been speeded up by a factor of 15-25, and the addition of a new interface for Correlation Spectroscopy. We’ve also added a lot of new file import/export options and further refined many of the plotting tools, making it easier than ever to get the information you want to see right in front of you.
Though considerable, the updates for PLS_Toolbox and Solo might be called evolutionary. But the MIA_Toolbox/Solo+MIA upgrade is revolutionary. The main interface for MIA is now the Image Manager, which provides a place to load, organize, explore, and manipulate images before further analysis with other Eigenvector tools. This plus the new Trend Tool make it easy to explore and edit multivariate images. The whole work flow is streamlined. Add this to the host of analysis methods available, the improved computational tools for things like MCR, and the availability of our tools for 64-bit platforms, and you’ve got a very powerful set of tools for dealing with large multivariate images!
Existing users with current maintenance contracts can down download the new tools from their accounts. New users can order or get free 30-day demos by creating an account.
Enjoy the updates!
BMW
Different kinds of PLS weights, loadings, and what to look at?
Sep 12, 2009
The age of Partial Least Squares (PLS) regression (as opposed to PLS path modeling) began with the SIAM publication of Svante Wold et. al. in 1984 [1]. Many of us learned PLS regression from the rather more accessible paper by Geladi and Kowalski from 1986 [2] which described the Nonlinear Iterative PArtial Least Squares (NIPALS) algorithm in detail.
For univariate y, the NIPALS algorithm is really more sequential than iterative. There is a specific sequence of fixed steps that are followed to find the weight vector w (generally normalized to unit length) for each PLS factor or Latent Variable (LV). Only in the case of multivariate Y is the algorithm really iterative, a sequence of steps is repeated until the solution for w for each LV converges.
Whether for univariate y or multivariate Y the calculations for each LV end with a deflation step. The X data is projected onto the weight vector w to get a score vector, t (t = Xw). X is then projected onto the score t to get a loading, p, (p = X’t/t’t). Finally, X is deflated by tp’ to form a new X, Xnew with which to start the procedure again, Xnew = X – tp’. A new weight vector w is calculated from the deflated Xnew and the calculations continue.
So the somewhat odd thing about the weight vectors w derived from NIPALS is that each one applies to a different X, i.e. tn+1 = Xnwn+1. This is in contrast to Sijmen de Jong’s SIMPLS algorithm introduced in 1993 [3]. In SIMPLS a set of weights, sometimes referred to as R, is calculated, which operate on the original X data to calculate the scores. Thus, all the scores T can be calculated directly from X without deflation, T = XR. de Jong showed that it is easy to calculate the SIMPLS R from the NIPALS W and P, R = W(P’W)-1. (Unfortunately, I have, as yet, been unable to come up with a simple expression for calculating the NIPALS W from the SIMPLS model parameters.)
So the question here is, “If you want to look at weights, which weights should you look at, W or R?” I’d argue that R is somewhat more intuitive as it applies to the original X data. Beyond that, if you are trying to standardize outputs of different software routines (which is actually how I got started on all this), it is a simple matter to always provide R. Fortunately, R and W are typically not that different, and in fact, they start out the same, w1 = r1, and they span the same subspace. Our decision here, based on relevance and standardization, is to present R weights as the default in future versions of PLS_Toolbox and Solo, regardless of which PLS algorithm is selected (NIPALS, SIMPLS or the new Direct Scores PLS).
A better question might be, “When investigating a PLS model, should I look at weights, R or W, or loadings P?” If your perspective is that the scores T are measures of some underlying “latent” phenomena, then you would choose to look at P, the degree to which these latent variables contribute to X. The weights W or R are merely regression coefficients that you use to estimate the scores T. From this viewpoint the real model is X = TP’ + E and y = Tb+ f.
If, on the other hand, you see PLS as simply a method for identifying a subspace within which to restrict, and therefore stabilize, the regression vector, then you would choose to look at the weights W or R. From this viewpoint the real model is Y = Xb + e, with b = W(P’W)-1(T’T)-1T’y = R(T’T)-1T’y via the NIPALS and SIMPLS formulations respectively. The regression vector is a linear combination of the weights W or R and from this perspective the loadings P are really a red herring. In the NIPALS formulation they are a patch left over from the way the weights were derived from deflated X. And the loadings P aren’t even in the SIMPLS formulation.
So the bottom line here is that you’d look at loadings P if you are a fan of the latent variable perspective. If you’re a fan of the regression subspace perspective, then you’d look at weights, W or preferably R. I’m in the former camp, (for more reasons than just philosophical agreement with the LV model), as evidenced by my participation in S. Wold et. al., “The PLS model space revisited,” [4]. Your choice of perspective also impacts what residuals to monitor, etc., but I’ll save that for a later time.
BMW
[1] S. Wold, A. Ruhe, H. Wold, and W.J. Dunn III, “The Collinearity Problem in Linear
Regression. The Partial Least Square Approach to Generalized Inverses”, SIAM J. Sci.
Stat. Comput., Vol. 5, 735-743, 1984.
[2] P. Geladi and B.R. Kowalski, “PLS Tutorial,” Anal. Chim. Acta., 185(1), 1986.
[3] S. de Jong, “SIMPLS: an alternative approach to partial least squares regression,” Chemo. and Intell. Lab. Sys., Vol. 18, 251-263, 1993.
[4] S. Wold, M. Høy, H. Martens, J. Trygg, F. Westad, J. MacGregor and B.M. Wise, “The PLS model space revisited,” J. Chemometrics, pps 67-68, Vol. 23, No. 2, 2009.
New software releases coming in October
Sep 11, 2009
The Eigenvector programming staff has been working hard all summer, and the fruits of their labor are just about ripe! In October we plan to release new and substantially improved versions of PLS_Toolbox, MIA_Toolbox, Solo, and Solo+MIA. For a rundown of the most significant improvements see the new features brochure.
I’m especially excited about the improvements to our Multivariate Image Analysis (MIA) tools. Our MATLAB-based MIA_Toolbox has undergone it’s most extensive upgrade since it’s introduction. It now features a new interface for organizing, editing and viewing images. There is also a new Texture Analysis interface, and lots of new image manipulation and filtering tools.
Our stand-alone Solo+MIA has all the features of the new MIA_Toolbox (of course!). Plus, we’re making Solo+MIA available for 64-bit operating systems. This makes working with the often very large images produced by advanced analytical techniques much more efficient.
We plan to release our new versions after the FACSS meeting, October 18-22. If you’d like to see them before then, visit us next week at SIMS XVII in Toronto, or at FACSS 2009 in Louisville. Better yet, take one of our hands-on courses at FACSS, where we’ll be using our new versions.
The best chemometrics software just keeps getting better!
BMW
Donal O’Sullivan joins Eigenvector
Sep 5, 2009
We’re pleased to announce that Donal O’Sullivan is the newest Eigenvectorian. As Senior Software Developer, Donal will be working with our Jeremy Shaver and Scott Koch to accelerate development of our off-the-shelf software products (PLS_Toolbox, Solo, etc.) and create custom solutions for our consulting clients.
Donal has a wealth of programming experience and a very interesting scientific background as well. After receiving his M.S. in Applied Mathematics at University College Dublin, he came to University of Washington where he completed his Ph.D. in Atmospheric Sciences. This was followed by several research positions focused on the computationally intensive problem of modeling atmospheric phenomena. Most recently, Donal has worked with biosoftware, in the Algorithm Development Group for the Rosetta Resolver and Syllego genetic data analysis systems.
We’re excited about working with Donal because, in addition to great programming skills, he brings experience working with a diverse set of applications. Beyond that, we find him to be quite thoughtful, dedicated to quality, and easy to be around!
Welcome Donal!
BMW
Back to School Deals for Chemometrics Novices
Aug 26, 2009
NOTE: Current pricing can be found here.
That nip in the air in the morning means that back to school time has arrived. Kids of all ages are getting their supplies together, and so are their teachers.
If you are a student or instructor in a chemometrics or related class, we’ve got a deal for you! We’ll let you use demo versions of any of our data modeling software packages free-of-charge for six months. This includes our stand-alone products Solo and Solo+MIA, and our MATLAB toolboxes PLS_Toolbox, MIA_Toolbox and EMSC_Toolbox. All our demos are fully functional (no data size limitations), include all documentation and lots of example data sets.
To get started with the program, just write to me with information about the class our software will be used with (course title, instructor, start and end dates). If professors send their class list, we’ll set up accounts for all the students allowing them access to the extended demos. Alternately, students can set up their own accounts and let us know what class they are taking.
Many professors have gravitated towards using our Solo products. They don’t require MATLAB, so students can just download them and they’re ready to go. Solo and Solo+MIA work on Windows (32 and 64 bit) and Mac OS X.
BMW
Referencing Software – Summary
Jun 27, 2009
The post I wrote on June 11, Referencing Software, resulted in a rather lengthy thread on ICS-L, the Chemometrics discussion list. Most discussants generally agreed that the software used to develop results for scientific papers should be referenced, including the software title, publisher, year and version number.
There were a few dissenters. Sergey Kucheryavski wrote:
…it makes sense only in two cases.
1. The software implements some unique, patented methods that are not available in any other programs.
2. There is a special agreement (caused by special price [e.g. PLS toolbox for 100 USD] or some other benefits) between user and a software company.
In a similar vein, Richard Brereton wrote:
If however software has been bought (often at considerable cost) I do not think this obliges the originator to cite it, there is no ethical requirement, unless the software was purchased at a specially low price…
I find this to be a rather interesting concept and wonder what would happen if we applied it to books as well. If I paid the $1595 for Comprehensive Chemometrics by Brown, Tauler and Walczak, do I not have to reference it? How about Brereton’s “Applied Chemometrics for Scientists?” Do I have to reference it because it’s only $110? Or do I have to reference it only if I get a discount off the list price? Clearly this is ridiculous.
Most of the respondents felt that it was important to reference software in order to assure reproducibility of the work. Philip Hopke wrote:
It seems to me that the fundamental issue is the ability of the reader of a paper to be able to fully replicate the work that has been published. If work is not replicable, then it is not science. The issue then is what is required to ensure repeatability of the … work being reported.
And I agree that this is the most important issue, but it is not the only issue. Bruce Kowalski wrote:
If ya’ll want a real debate start taking issue with publications that don’t mention the first paper in a research area. There has always been a correct way to use references. What went wrong???????
Referencing the first paper in a research area is all about giving credit where it’s due. It’s not about reproducibility, (which, as Cliff Spiegelman observed, is often better served by referencing later work, often done by someone other than the original author). Likewise, referencing software is also partly about giving credit where it’s due–recognizing the effort expended to produce a useful tool of scientific value.
BMW
Software Engineer Sought
Jun 19, 2009
The current recession hasn’t slowed things down here at Eigenvector. In fact, we’re looking for another Software Engineer to help us speed up evolution of our current products and develop custom solutions for our consulting clients. Please find below an ad for the position.
BMW
Software Engineer sought for leading role in scientific software development.
Job Description:
The successful candidate will work with our Seattle-based scientific staff to develop MATLAB-based and stand-alone chemometrics products, including our flagship PLS_Toolbox and Solo. This includes design and implementation of interfaces and supporting documentation and development and refinement of numerical algorithms. Will also develop custom solutions for clients, including on-line applications.
About Eigenvector Research:
Eigenvector Research is a chemometrics research and applications company with headquarters in Wenatchee, Washington. Chemometrics is the application of mathematical and statistical methods to problems in chemistry. Our PLS_Toolbox product is used by engineers and scientists in 50+ countries. We also provide training and consulting services for a wide variety of clients in the chemical, petrochemical, consumer products, food & agriculture, pharmaceutical, semiconductor and medical device fields. For more information, please visit our website.
Position Requirements:
Must have experience with MATLAB and either C/C++ or Java, and be familiar with object-oriented programming techniques and development of graphical user interfaces. Strong math skills, especially linear algebra, desired. Background in multivariate statistics and chemical sciences a plus. Good communication skills, both oral and written, required. Must be self-starter, willing to work at home, and productive with only moderate supervision. B.S. or higher degree in chemistry, chemical engineering or other relevant scientific discipline preferred. Three years or more experience desired. Close proximity to Seattle a plus.
To Apply:
Send your cover letter and resume to Barry M. Wise, bmw@eigenvector.com.
Referencing Software
Jun 11, 2009
Yesterday I picked up a newly-arrived journal and noted that the article highlighted on the front page looked quite interesting as we have been doing some related work. I eagerly turned to the article and found that the author had been using PLS-DA (Partial Least Squares Discriminant Analysis) and was, in fact, one of our PLS_Toolbox users. Imagine my disappointment when I could find no reference to our software in the article! The article had many references, with more than 50 journal articles cited and at least a half dozen equipment manufacturers named. But there was no mention of the software that was used to turn the measurements into the results that were presented.
I checked with the author of the article, and yes, our software was used to produce the results that were re-plotted in another application prior to publication. But exactly whose software was used is beside the point. The point is that software is a critical part of the experiment and, in order to ensure reproducibility, should be referenced.
Some might say that referencing the original work on the development of the particular analysis method implemented in the software should suffice, (though in this instance that wasn’t done either, the author referenced previous work of their own where they used PLS-DA). I’d argue that isn’t enough. The problem is that it typically takes a LOT of additional work to turn a method from an academic paper into a working program. There are often many special (sometimes schizophrenic) cases that must be handled properly to assure consistent results. Sometimes various meta-parameters must be optimized. Preprocessing can be critical. And then there is the whole interface which allows the user to interact with the data so that it can be effectively viewed, sorted and selected.
So why do I care? Obviously, there is the commercial aspect: having our software referenced is good advertising for us, and leads to more sales. But beyond that, (like many other publishers of scientific software, I’m sure), our software is probably our most significant work of scholarship. To not reference it is to not acknowledge the contributions we’ve made to the field.
So I’m calling on all authors to reference the software they use, and editors and reviewers to check that they do. Listing it in the references would be preferred. Software is, after all, a publication, with a title, version number (edition), publisher, and year of publication. Often, authors are also known, and can be listed. But one way or the other, software should be cited as it is critical to reproducibility and represents scholarly work upon which results depend. Referencing software upholds scientific and scholarly tradition and it is academically dishonest to not do so.
BMW

 SEARCH
SEARCH